Everyone who studies molecular biology, biochemistry, genetic engineering, and a number of other related sciences, sooner or later asks the question: what function does RNA polymerase perform? This is a rather complicated topic, which has not yet been fully explored, but, nevertheless, what is known will be covered in the article.
general information
It must be remembered that there is RNA polymerase of eukaryotes and prokaryotes. The first is further divided into three types, each of which is responsible for the transcription of a separate group of genes. These enzymes are numbered for simplicity as the first, second, and third RNA polymerase.
Prokaryote, whose
structure is nuclear-free, during transcription acts according to a simplified scheme. Therefore, for clarity, to cover as much information as possible, eukaryotes will be considered. RNA polymerases are structurally similar. It is believed that they contain at least 10 polypeptide chains. In this case, RNA polymerase 1 synthesizes (transcribes) genes, which will subsequently be translated into various proteins. The second deals with the transcription of genes, which are subsequently translated into proteins. RNA polymerase 3 is represented by a variety of low molecular weight stable enzymes that are moderately sensitive to alpha-amatin. But we have not decided what RNA polymerase is! So called enzymes that are involved in the synthesis of
ribonucleic acid molecules
. In a narrow sense, this refers to DNA-dependent RNA polymerases that act on the basis of a
deoxyribonucleic acid matrix
. Enzymes are of utmost importance for the long and successful functioning of living organisms. RNA polymerase can be found in all cells and most viruses.
Feature Division
Depending on the subunit composition of RNA polymerase are divided into two groups:
- The first is transcribing a small number of genes in simple genomes. For functioning in this case, complex regulatory influences are not required. Therefore, this includes all enzymes that consist of only one subunit. As an example, RNA polymerase of bacteriophages and mitochondria can be induced.
- This group includes all RNA polymerases of eukaryotes and bacteria that are complex in structure. They are intricate multisubunit protein complexes that can transcribe thousands of different genes. During functioning, these genes respond to a large number of regulatory signals that come from protein factors and nucleotides.
Such structural and functional separation is a very conditional and strong simplification of the real situation.
What does RNA polymerase I do?
They are assigned the function of the formation of primary transcripts of rRNA genes, that is, they are the most important. The latter are better known under the designation 45S-RNA. Their length is approximately 13 thousand nucleotides. 28S-RNA, 18S-RNA and 5,8S-RNA are formed from it. Due to the fact that only one transcriptor is used to create them, the body receives a “guarantee” that the molecules will form in equal amounts. In this case, only 7 thousand nucleotides are used to create RNA directly. The rest of the transcript is degraded in the nucleus. Regarding such a large residue, there is an opinion that it is necessary for the early stages of ribosome formation. The number of these polymerases in the cells of higher beings fluctuates around the mark of 40 thousand units.
How is it organized?
So, we have already considered quite well the first RNA polymerase (prokaryotic structure of a molecule). Moreover, large subunits, as well as a large number of other high molecular weight polypeptides, there are well distinguishable functional and structural domains. During the cloning of genes and determination of their primary structure, scientists have identified evolutionarily conservative sections of chains. Using good expression, the researchers also conducted a mutational analysis, which allows us to talk about the functional significance of individual domains. To do this, using directed mutagenesis in polypeptide chains, individual amino acids were changed and such altered subunits were used in the assembly of enzymes, followed by analysis of the properties that were obtained in these constructs. It was noted that due to its organization, the first RNA polymerase does not react in any way to the presence of alpha-amatin (a highly toxic substance that is obtained from pale grebe).
Functioning
Both the first and second RNA polymerases can exist in two forms. One of them may act to initiate specific transcription. The second is DNA dependent
RNA polymerase. This ratio is manifested in the magnitude of the activity of functioning. The topic is still being investigated, but it is already known that this depends on two transcription factors, which are designated as SL1 and UBF. A feature of the latter is that it can directly bind to the promoter, whereas SL1 requires the presence of UBF. Although it was experimentally established that DNA-dependent RNA polymerase can participate in transcription at a minimum level and without the presence of the latter. But for the normal functioning of this mechanism, UBF is still needed. Why so? Reliably so far it has not been possible to establish the reason for this behavior. One of the most popular explanations suggests that UBF acts as a kind of stimulator of rDNA transcription when it grows and develops. When the dormant phase sets in, the minimum necessary level of functioning is maintained. And for him, the participation of transcription factors is not critical. This is how
RNA polymerase works
. The functions of this enzyme make it possible to support the reproduction of small “building blocks” of our body, thanks to which it is constantly updated for decades.
The second group of enzymes
Their functioning is regulated by the assembly of a multi-protein pre-initiator complex of second-class promoters. Most often this is expressed in the work with special proteins - activators. An example is TBP. These are the associated factors that make up the TFIID. They are the target for p53, NF kappa B and so on. Proteins called coactivators also exert their influence in the regulation process. An example is GCN5. Why are these proteins needed? They act as adapters that configure the interaction of activators and factors that are part of the pre-initiator complex. For transcription to occur correctly, the necessary initiating factors are necessary. Despite the fact that there are six of them, only one can directly interact with the promoter. For other cases, a preformed complex of the second RNA polymerase is required. Moreover, during these processes, the proximal elements are nearby - only 50-200 pairs from the site where transcription began. They contain an indication of the binding of activator proteins.
Specific features
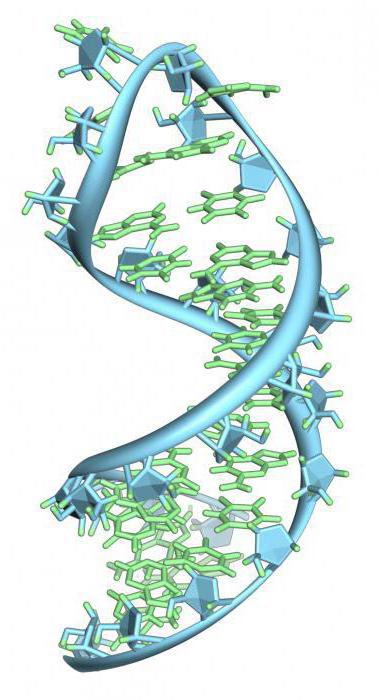
Does the subunit structure of enzymes of different origin affect their functional role in transcription? There is no exact answer to this question, but it is believed that it is most likely positive. How does RNA polymerase depend on this? The functions of simple-structure enzymes are the transcription of a limited circle of genes (or even their small parts). An example is the synthesis of RNA seeds of Okazaki fragments. The promoter specificity of bacterial and phage RNA polymerase lies in the fact that enzymes possess a simple structure and do not differ in variety. This can be seen in the example of DNA replication in bacteria. Although one can consider the following: when the complex structure of the even T-phage genome was studied, during the development of which multiple switching of transcription between different groups of genes was noted, it was revealed that a complex host RNA polymerase was used for this. That is, a simple enzyme is not induced in such cases. A number of consequences follow from this:
- RNA polymerase of eukaryotes and bacteria must be able to recognize different promoters.
- It is necessary that the enzymes have a certain reaction to different regulatory proteins.
- RNA polymerase must also be able to change the specificity of recognition of the nucleotide sequence of template DNA. For this, a variety of protein effectors are used.
From here follows the body's need for additional “building” elements. Proteins of the transcribing complex help RNA polymerase to fully perform their functions. This applies, to the greatest extent, to enzymes of complex structure, in the capabilities of which the implementation of an extensive program for the realization of genetic information. Due to various tasks, we can observe a peculiar hierarchy of the structure of RNA polymerases.
How does the transcription process go?
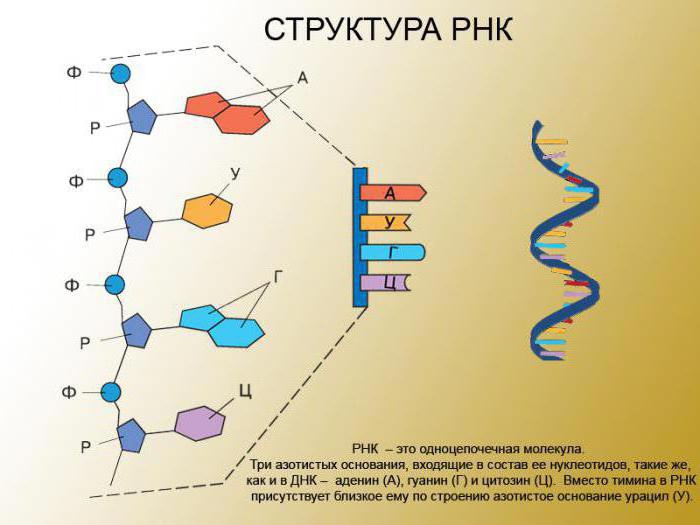
Is there a gene responsible for binding to RNA polymerase? To begin with, transcription: in eukaryotes, the process occurs in the nucleus. In prokaryotes, it flows inside the microorganism itself. The interaction of polymerase is based on the fundamental structural principle of the complementary pairing of individual molecules. Regarding the issues of interaction, we can say that DNA acts exclusively as a matrix and does not change during transcription. Since DNA is an integral enzyme, it is certain to say that a particular gene is responsible for this polymer, but it will take a very long time. It should not be forgotten that DNA contains 3.1 billion nucleotide residues. Therefore, it will be more appropriate to say that each type of RNA has its own DNA. For the polymerase reaction to take place, energy sources and ribonucleoside triphosphato substrates are needed. If they are present, 3 ', 5'-phosphodiester bonds between ribonucleoside monophosphates are formed. The RNA molecule begins to be synthesized in certain DNA sequences (promoters). This process ends at the termination sites (termination). The site that is involved here is called a transcripton. Here, as a rule, eukaryotes have only one gene, while prokaryotes can possess several sections of the code. Each transcripton has a non-informative zone. They contain specific nucleotide sequences that interact with regulatory transcription factors mentioned earlier.
Bacterial RNA Polymerases
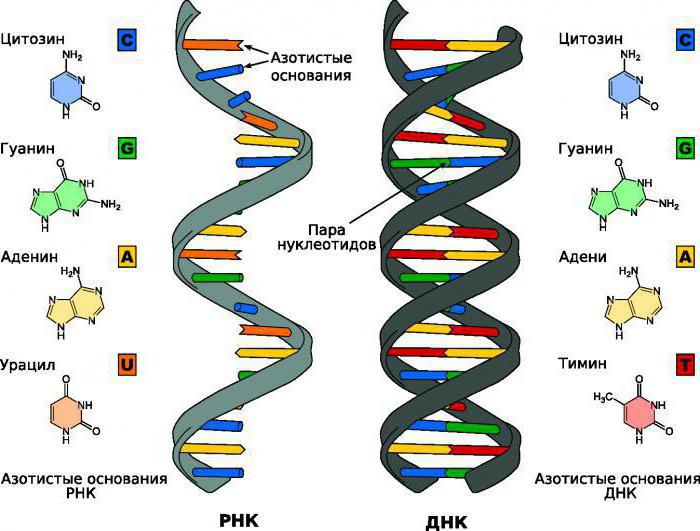
In these microorganisms, one enzyme is responsible for the synthesis of mRNA, rRNA and tRNA. The average polymerase molecule has about 5 subunits. Two of them act as binding elements of the enzyme. Another subunit is involved in the initiation of synthesis. There is also an enzyme component for non-specific binding to DNA. And the last subunit is engaged in bringing the RNA polymerase into working form. It should be noted that the enzyme molecules are not in "free" swimming in the cytoplasm of the bacterium. When RNA polymerases are not used, they bind to non-specific regions of DNA and wait for the active promoter to open. A little distracting from the topic, it should be said that it is very convenient to study proteins and their effect on ribonucleic acid polymerases on bacteria. It is especially convenient to put experiments on stimulation or inhibition of individual elements on them. Due to their high breeding rate, the desired result can be obtained relatively quickly. Alas, human research cannot be carried out at such a fast pace due to our structural diversity.
How did RNA polymerase "take root" in various forms?
So the article comes to a logical conclusion. The focus was on eukaryotes. But there are still archaea and viruses. Therefore, I want to pay a little attention to these forms of life. In the life of archaea there is only one group of RNA polymerases. But it is extremely similar in its properties to the three associations of eukaryotes. Many scholars have suggested that what we can observe in archaea is actually the evolutionary ancestor of specialized polymerases. The structure of viruses is also interesting. As previously written, not all such microorganisms have their own polymerase. And where it is, it is one subunit. It is believed that viral enzymes come from DNA polymerases, and not from complex RNA constructs. Although, due to the diversity of this group of microorganisms, there is a different implementation of the biological mechanism under consideration.
Conclusion
Alas, humanity does not yet have all the necessary information necessary for understanding the genome. And what could only be done! Almost all diseases basically have a genetic basis - this applies primarily to viruses that constantly cause us problems, infections, and so on. The most complex and incurable diseases - they also, in fact, directly or indirectly depend on the human genome. When we learn to understand ourselves and will apply this knowledge to the benefit, a large number of problems and diseases will simply cease to exist. Already now many previously terrible diseases, such as smallpox and plague, have receded into the past. Pig and whooping cough are preparing to go there. But we should not relax, because we still have a large number of different challenges that need to be answered. And he will be found, for everything goes to this.